Select The Repetitive Dna Element From The Choices Below.
Holbox
Mar 24, 2025 · 6 min read
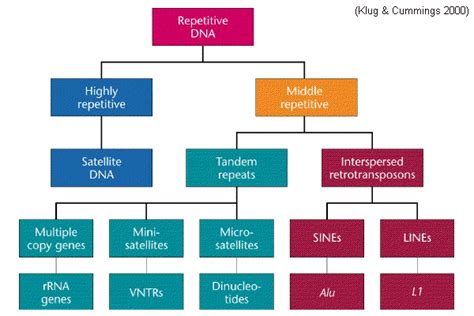
Table of Contents
- Select The Repetitive Dna Element From The Choices Below.
- Table of Contents
- Selecting Repetitive DNA Elements: A Deep Dive into Genome Architecture
- Defining Repetitive DNA
- Distinguishing Features of Repetitive DNA
- Major Classes of Repetitive DNA
- 1. Tandem Repeats: Repeated Sequences in a Row
- 2. Interspersed Repeats: Scattered Throughout the Genome
- The Significance of Repetitive DNA
- Identifying and Analyzing Repetitive DNA
- Conclusion: The Undervalued Complexity of Repetitive DNA
- Latest Posts
- Latest Posts
- Related Post
Selecting Repetitive DNA Elements: A Deep Dive into Genome Architecture
Repetitive DNA elements are a significant component of eukaryotic genomes, playing crucial, albeit often enigmatic, roles in genome structure, function, and evolution. Understanding these elements is key to comprehending the complexities of genetic inheritance, genomic instability, and the evolution of species. This article explores the diverse world of repetitive DNA, examining various types and highlighting their importance in biological processes. We'll also delve into methods used for their identification and analysis.
Defining Repetitive DNA
Repetitive DNA, as the name suggests, comprises sequences that are repeated multiple times throughout the genome. These repeats can range from short, simple sequences to long, complex units, and they can be dispersed throughout the genome or clustered in specific regions. The sheer abundance of repetitive DNA distinguishes eukaryotic genomes from prokaryotic ones, which generally have far less repetitive content. The proportion of repetitive DNA varies considerably between species, even within closely related organisms.
Distinguishing Features of Repetitive DNA
Several key features help categorize and differentiate repetitive DNA elements:
- Sequence Length: This can range from a few base pairs (like microsatellites) to hundreds of kilobases (like transposable elements).
- Copy Number: The number of times a specific sequence is repeated varies dramatically across different repetitive elements.
- Sequence Homology: The degree of similarity between different copies of a repeat. Some are nearly identical, while others may show substantial divergence due to mutations accumulated over time.
- Chromosomal Distribution: Repeats can be dispersed across chromosomes (interspersed repeats) or clustered in specific locations, such as centromeres and telomeres (tandem repeats).
- Transcriptional Activity: Some repetitive elements are actively transcribed, while others are largely silent. The transcripts produced can have regulatory functions or contribute to non-coding RNA populations.
Major Classes of Repetitive DNA
Repetitive DNA encompasses a vast array of elements, broadly classified into several major categories:
1. Tandem Repeats: Repeated Sequences in a Row
Tandem repeats are characterized by consecutive, identical or nearly identical sequences arranged head-to-tail. They're often found in specific chromosomal locations and are broadly categorized based on their repeat unit length:
-
Satellite DNA: These are long arrays of short, highly repetitive sequences. They are typically found in centromeres and heterochromatin, playing a critical role in chromosome segregation during cell division. Different types of satellite DNA are classified based on their buoyant density, revealed through density gradient centrifugation.
-
Minisatellites (Variable Number Tandem Repeats - VNTRs): These have longer repeat units (10-100 bp) and are highly polymorphic, meaning they exhibit extensive variation between individuals. This polymorphism makes them valuable tools in forensic science and paternity testing.
-
Microsatellites (Short Tandem Repeats - STRs): These are the shortest tandem repeats (1-6 bp) and exhibit extremely high variability between individuals. Their high mutation rate makes them particularly useful in population genetics studies and for tracing ancestry.
2. Interspersed Repeats: Scattered Throughout the Genome
Interspersed repeats are scattered throughout the genome rather than clustered in specific regions. These are often mobile genetic elements, capable of transposition to new genomic locations. The most prominent class of interspersed repeats are transposable elements (TEs), also known as transposons or "jumping genes."
-
Transposable Elements (TEs): These are DNA sequences capable of moving from one genomic location to another. TEs can be further classified into two main types:
-
DNA Transposons: These move directly through a "cut-and-paste" mechanism, excising themselves from their original location and inserting into a new site.
-
Retrotransposons: These transpose via an RNA intermediate. They are transcribed into RNA, reverse-transcribed into DNA, and then integrated into a new genomic location. Retrotransposons are further sub-categorized into different families, including long terminal repeat (LTR) retrotransposons and non-LTR retrotransposons (like LINEs and SINEs). Long interspersed nuclear elements (LINEs) are long sequences that can self-replicate. Short interspersed nuclear elements (SINEs) are shorter and require LINE-encoded machinery for their transposition.
-
The Significance of Repetitive DNA
Despite their often repetitive nature, these elements are far from inert. Their importance in various biological processes is becoming increasingly apparent:
-
Genome Structure and Organization: Repetitive DNA plays a fundamental role in shaping the three-dimensional structure of chromosomes. Satellite DNA, for instance, is crucial for centromere function and chromosome segregation.
-
Gene Regulation: Some repetitive elements act as regulatory sequences, influencing the expression of nearby genes. Others may contribute to the formation of heterochromatin, which is generally transcriptionally inactive.
-
Genomic Evolution: Repetitive DNA is a major driver of genomic evolution. Transposable elements can contribute to gene duplication, exon shuffling, and the creation of new genes. They can also cause mutations by inserting into genes or disrupting regulatory sequences. However, their effects are not solely disruptive; some TEs have been co-opted by the host genome to serve beneficial functions.
-
Disease Association: Aberrations in repetitive DNA can be associated with several human diseases. Expansions of trinucleotide repeats, for example, are responsible for a range of neurological disorders, including Huntington's disease and fragile X syndrome. Changes in copy number of certain repetitive elements can also contribute to cancer development.
-
Forensic Science and Ancestry Studies: The high polymorphism of microsatellites and minisatellites makes them invaluable tools in forensic science and paternity testing. Their variability also allows researchers to trace human migration patterns and understand population history.
Identifying and Analyzing Repetitive DNA
Identifying and analyzing repetitive DNA requires specialized techniques:
-
DNA Sequencing: High-throughput sequencing technologies have revolutionized our ability to identify and characterize repetitive DNA. Sequencing allows researchers to determine the exact sequences and copy numbers of repetitive elements.
-
Bioinformatics: Powerful bioinformatics tools are essential for analyzing large sequencing datasets. These tools can help identify repetitive elements, assess their sequence similarity, and predict their potential function.
-
Fluorescence In Situ Hybridization (FISH): FISH is a cytogenetic technique used to visualize repetitive DNA sequences on chromosomes. Fluorescently labeled probes are designed to hybridize with specific repetitive sequences, allowing researchers to determine their chromosomal location and abundance.
-
Southern Blotting: This technique can be used to detect the presence and size of repetitive elements in a genome. It involves digesting genomic DNA with restriction enzymes, separating the fragments by gel electrophoresis, and then probing the gel with a labeled probe complementary to the repetitive sequence.
Conclusion: The Undervalued Complexity of Repetitive DNA
Repetitive DNA elements, far from being mere genomic "junk," are integral components of eukaryotic genomes. They contribute substantially to genome architecture, gene regulation, and evolution. While their functions remain only partially understood, ongoing research using advanced sequencing and bioinformatics techniques continue to illuminate their roles in shaping life as we know it. Understanding these elements is critical not only for basic biological research but also for advancing medical diagnostics, forensic science, and our understanding of human evolution and disease. The continued exploration of repetitive DNA promises further insights into the intricacies of the genome and its profound impact on life's diversity and complexity.
Latest Posts
Latest Posts
-
When Supplies Are Purchased On Credit It Means That
Mar 26, 2025
-
P Is The Insured On A Participating Life Policy
Mar 26, 2025
-
On July 1 A Company Receives An Invoice
Mar 26, 2025
-
The Prokaryotic Cells That Built Stromatolites Are Classified As
Mar 26, 2025
-
A Nation Can Achieve Higher Economic Growth If
Mar 26, 2025
Related Post
Thank you for visiting our website which covers about Select The Repetitive Dna Element From The Choices Below. . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
