The Nucleic Acid Sequence In Mrna Is Determined By
Holbox
Mar 16, 2025 · 7 min read
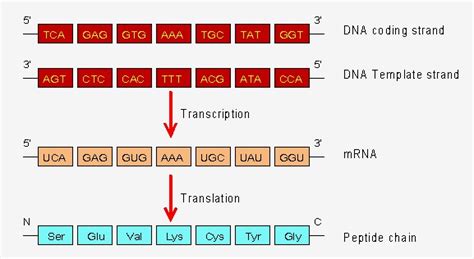
Table of Contents
The Nucleic Acid Sequence in mRNA is Determined By: A Deep Dive into Transcription and its Regulation
The central dogma of molecular biology dictates that information flows from DNA to RNA to protein. A crucial step in this process is the transcription of DNA into messenger RNA (mRNA), where the nucleic acid sequence of the mRNA molecule is faithfully (though not always perfectly) copied from the DNA template. Understanding how this sequence is determined is fundamental to comprehending gene expression, regulation, and the myriad biological processes they underpin. This article will delve into the intricate mechanisms that govern this critical step, exploring the enzymes, regulatory elements, and cellular processes involved.
The Transcription Machinery: Enzymes and Factors
The process of transcription is orchestrated by a complex molecular machine, primarily involving RNA polymerase and a suite of transcription factors. Let's break down the key players:
RNA Polymerase: The Transcription Enzyme
RNA polymerase is the central enzyme responsible for synthesizing the mRNA molecule. Unlike DNA polymerase, which requires a primer to initiate synthesis, RNA polymerase can initiate transcription de novo. In eukaryotes, three distinct RNA polymerases exist:
- RNA Polymerase I: Primarily transcribes ribosomal RNA (rRNA) genes.
- RNA Polymerase II: Transcribes protein-coding genes, producing mRNA. This is the polymerase we will focus on for the context of mRNA sequence determination.
- RNA Polymerase III: Transcribes transfer RNA (tRNA) genes and other small RNA molecules.
Transcription Factors: The Regulators
Transcription factors are proteins that bind to specific DNA sequences, influencing the initiation and rate of transcription. They are crucial for determining which genes are transcribed and at what level. These factors can be broadly categorized as:
-
General Transcription Factors (GTFs): These are essential for the basic transcription machinery to function. They assemble at the promoter region of a gene, forming the pre-initiation complex (PIC) which recruits RNA polymerase II. In eukaryotes, key GTFs include TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH.
-
Specific Transcription Factors (STFs): Also known as regulatory transcription factors, these proteins bind to specific DNA sequences called enhancer or silencer elements, located either upstream, downstream, or even within the gene. They modulate the activity of the general transcription factors, either increasing (activators) or decreasing (repressors) the rate of transcription. This level of control allows cells to precisely regulate gene expression in response to various stimuli and developmental cues.
The interaction between GTFs and STFs, along with other regulatory proteins, determines the overall transcription efficiency of a specific gene. The precise combination of transcription factors bound to a particular promoter region dictates the efficiency with which RNA polymerase initiates transcription, and therefore, fundamentally influences the mRNA sequence generated.
The DNA Template: Promoters, Enhancers, and Silencers
The DNA sequence itself plays a crucial role in determining the mRNA sequence. Specific DNA elements guide the binding of RNA polymerase and transcription factors, dictating where and when transcription begins and ends. These include:
Promoters: The Starting Point
Promoters are DNA sequences located upstream of the transcription start site (TSS). They contain specific consensus sequences, such as the TATA box in eukaryotes, that serve as binding sites for the general transcription factors and RNA polymerase II. The promoter's strength, determined by its sequence and the number of binding sites for transcription factors, dictates the basal level of transcription. Variations in the promoter sequence can significantly impact the amount of mRNA produced.
Enhancers: The Accelerators
Enhancers are DNA regulatory elements that can increase the rate of transcription, even when located far from the promoter region, either upstream or downstream. They act as binding sites for specific transcription factors known as activators, which interact with the general transcription machinery at the promoter, enhancing the initiation process. The distance between enhancers and promoters is not a limiting factor; the DNA looping allows for physical proximity and interaction between the bound activators and the transcription machinery.
Silencers: The Brakes
Silencers, analogous to enhancers, are DNA sequences that negatively regulate transcription by binding repressor proteins. These repressors can interfere with the assembly of the PIC, or they may actively block the progression of RNA polymerase along the DNA template. Silencers contribute to the precise control of gene expression, ensuring that genes are only active when and where they are needed.
The Transcription Process: From DNA to mRNA
The process of transcription can be divided into three main phases:
Initiation: Assembling the Machinery
This phase involves the assembly of the PIC at the promoter. General transcription factors bind to the promoter region, followed by the recruitment of RNA polymerase II. Specific transcription factors bound to enhancers or silencers interact with the PIC, influencing the efficiency of initiation. The opening of the DNA double helix at the promoter forms a transcription bubble, allowing RNA polymerase access to the template strand.
Elongation: Synthesizing the mRNA
Once initiation is complete, RNA polymerase begins to synthesize the mRNA molecule. It does so by moving along the template DNA strand in the 3' to 5' direction, adding complementary ribonucleotides to the growing mRNA strand in the 5' to 3' direction. The precise sequence of nucleotides in the mRNA is dictated directly by the DNA template sequence. This phase involves the unwinding of the DNA helix ahead of the polymerase and the rewinding behind it.
Termination: Ending Transcription
Transcription termination is a complex process that varies among organisms and genes. In eukaryotes, the process involves the cleavage of the nascent mRNA molecule downstream of a polyadenylation signal sequence (AAUAAA). This cleavage is followed by the addition of a poly(A) tail to the 3' end of the mRNA molecule, which protects the mRNA from degradation. The remaining RNA is then further processed before being transported out of the nucleus.
Post-Transcriptional Modifications: Fine-Tuning the mRNA Sequence
The mRNA molecule produced during transcription undergoes several post-transcriptional modifications, further influencing the final sequence and its ability to be translated into a protein:
-
5' Capping: A 7-methylguanosine cap is added to the 5' end of the mRNA. This cap protects the mRNA from degradation and plays a role in the initiation of translation.
-
Splicing: Non-coding regions of the pre-mRNA called introns are removed, and the coding regions called exons are joined together. Alternative splicing allows a single gene to produce multiple mRNA isoforms with different sequences, expanding the proteome. The specific splicing pattern is influenced by various regulatory factors.
-
Polyadenylation: The addition of a poly(A) tail to the 3' end of the mRNA stabilizes the molecule and enhances its translation efficiency.
Errors and Variations: Imperfect Replication and Beyond
While the process of transcription strives for accuracy, errors can occur. These errors can lead to mutations in the mRNA sequence, potentially affecting the resulting protein. However, the cell has mechanisms to minimize these errors, including proofreading by RNA polymerase.
Additionally, variations in the mRNA sequence can arise through processes such as:
-
RNA editing: Chemical modifications to the mRNA sequence after transcription, altering its coding potential.
-
Transcriptional slippage: Errors in the alignment of DNA and RNA polymerase during elongation, leading to insertions or deletions in the mRNA sequence.
Conclusion: A Symphony of Molecular Interactions
The determination of the nucleic acid sequence in mRNA is a complex and tightly regulated process involving a multitude of molecular players. The DNA template, with its promoters, enhancers, and silencers, provides the blueprint, while RNA polymerase and transcription factors orchestrate the synthesis of the mRNA molecule. Post-transcriptional modifications further refine the sequence, influencing its stability and translational efficiency. A deep understanding of this process is essential for unraveling the intricacies of gene expression and its role in various biological phenomena, from development to disease. The interplay between these elements, along with the potential for error and variation, paints a dynamic picture of gene regulation, highlighting the elegance and complexity of life at the molecular level. Future research into these mechanisms will continue to reveal further insights into this fundamental aspect of cellular biology.
Latest Posts
Latest Posts
-
How Does A Shortcut Link To Another File
Mar 17, 2025
-
Cash Flows From Financing Activities Do Not Include
Mar 17, 2025
-
A Positive Return On Investment For Education Happens When
Mar 17, 2025
-
What Is The Value Of I
Mar 17, 2025
-
The Accounts In The Ledger Of Monroe Entertainment Co
Mar 17, 2025
Related Post
Thank you for visiting our website which covers about The Nucleic Acid Sequence In Mrna Is Determined By . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
