Choose All Modifications Of Eukaryotic Transcripts.
Holbox
Mar 25, 2025 · 6 min read
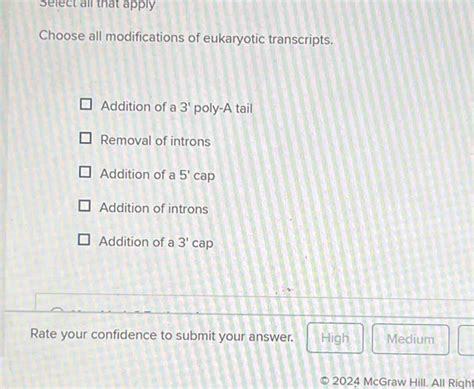
Table of Contents
- Choose All Modifications Of Eukaryotic Transcripts.
- Table of Contents
- Choosing All Modifications of Eukaryotic Transcripts: A Comprehensive Guide
- Beyond Transcription: The World of Post-Transcriptional Modifications
- 1. 5' Capping: Protecting and Guiding the Transcript
- 2. 3' Polyadenylation: Stability and Translation Control
- 3. RNA Splicing: Excising Introns and Joining Exons
- 4. RNA Editing: Altering the Nucleotide Sequence
- 5. RNA Methylation: Fine-Tuning Gene Expression
- 6. RNA Cleavage and Degradation: Quality Control and Gene Regulation
- The Interplay of Modifications: A Coordinated Effort
- The Biological Significance of Post-Transcriptional Modifications
- Technological Advances and Future Directions
- Latest Posts
- Latest Posts
- Related Post
Choosing All Modifications of Eukaryotic Transcripts: A Comprehensive Guide
Eukaryotic gene expression is a remarkably intricate process, far exceeding the simple transcription-translation paradigm of prokaryotes. A key element of this complexity lies in the extensive post-transcriptional modifications eukaryotic transcripts undergo before they can be translated into functional proteins. These modifications are crucial for regulating gene expression, ensuring mRNA stability, and ultimately determining the proteome's composition and functionality. This article will delve into the diverse range of modifications eukaryotic transcripts experience, exploring their mechanisms and biological significance.
Beyond Transcription: The World of Post-Transcriptional Modifications
The journey of a eukaryotic transcript from nascent RNA to mature mRNA is a carefully orchestrated sequence of events, far from a passive process. Several modifications act in concert to fine-tune gene expression at the post-transcriptional level. These modifications broadly fall under several categories:
1. 5' Capping: Protecting and Guiding the Transcript
The 5' cap is a crucial modification added to the 5' end of nascent pre-mRNA molecules. This cap consists of a 7-methylguanosine (m7G) residue linked to the first nucleotide via an unusual 5'-5' triphosphate linkage. This seemingly simple addition provides several critical functions:
-
Protection from Degradation: The 5' cap shields the mRNA from exonucleases, enzymes that degrade RNA from the 5' end, thus significantly enhancing mRNA stability.
-
Facilitating Translation Initiation: The cap structure is essential for the recruitment of the ribosome to the mRNA. Eukaryotic initiation factor 4E (eIF4E) binds specifically to the m7G cap, initiating the formation of the translation initiation complex.
-
Influencing Splicing and Nuclear Export: Evidence suggests the 5' cap plays a role in the efficiency of pre-mRNA splicing and its subsequent export from the nucleus to the cytoplasm, where translation occurs.
2. 3' Polyadenylation: Stability and Translation Control
The 3' end of eukaryotic transcripts undergoes polyadenylation, a process where a poly(A) tail – a long string of adenine nucleotides – is added. This process involves:
-
Cleavage: The pre-mRNA is cleaved at a specific site downstream of a polyadenylation signal sequence (typically AAUAAA).
-
Poly(A) Polymerase Activity: Poly(A) polymerase adds adenine residues to the 3' end of the cleaved transcript, creating the poly(A) tail. The length of this tail varies but is typically 200-250 nucleotides long.
The poly(A) tail plays a significant role in:
-
mRNA Stability: The poly(A) tail protects the mRNA from degradation by exonucleases. The length of the tail is dynamically regulated, affecting mRNA lifespan.
-
Translation Efficiency: The poly(A) tail, along with poly(A)-binding proteins (PABPs), interacts with translation initiation factors, enhancing translation initiation.
-
Nuclear Export: Polyadenylation is coupled with mRNA export from the nucleus.
3. RNA Splicing: Excising Introns and Joining Exons
Eukaryotic genes are composed of exons (coding sequences) and introns (non-coding intervening sequences). Pre-mRNA transcripts contain both exons and introns. RNA splicing is the process of removing introns and joining exons to generate a continuous coding sequence.
Splicing is catalyzed by the spliceosome, a large ribonucleoprotein complex composed of small nuclear RNAs (snRNAs) and proteins. The spliceosome recognizes specific sequences at the intron-exon boundaries (splice sites) and carries out two transesterification reactions to remove the intron and ligate the exons.
Alternative Splicing: A significant aspect of splicing is its ability to produce multiple mRNA isoforms from a single gene through alternative splicing. This process involves the differential inclusion or exclusion of exons during splicing, leading to the production of protein isoforms with different functions. Alternative splicing expands the proteome's diversity significantly.
4. RNA Editing: Altering the Nucleotide Sequence
RNA editing is a post-transcriptional process that alters the nucleotide sequence of the RNA molecule, leading to changes in the amino acid sequence of the translated protein. Several types of RNA editing exist, including:
-
Adenosine-to-inosine (A-to-I) Editing: Adenosine residues are deaminated to inosine, which is read as guanosine during translation.
-
Cytidine-to-uridine (C-to-U) Editing: Cytidine residues are deaminated to uridine, altering the codon sequence.
RNA editing can significantly affect protein function and regulation, often with significant physiological consequences.
5. RNA Methylation: Fine-Tuning Gene Expression
RNA methylation is the addition of a methyl group to a nucleotide base, most commonly adenine and cytosine. Methylation patterns are dynamic and can influence several aspects of RNA function, including:
-
Splicing: RNA methylation can affect splice site recognition and alternative splicing.
-
mRNA Stability: Methylation can affect mRNA stability by altering its interaction with RNA-binding proteins.
-
Translation Efficiency: Methylation patterns can influence the efficiency of translation initiation and elongation.
6. RNA Cleavage and Degradation: Quality Control and Gene Regulation
RNA degradation is a crucial process for controlling gene expression and maintaining the integrity of the transcriptome. Several pathways exist to degrade RNA molecules, including:
-
Exonuclease-mediated Degradation: Exonucleases degrade RNA from the 5' or 3' ends.
-
Endonuclease-mediated Degradation: Endonucleases cleave RNA at specific sites within the molecule.
-
Nonsense-mediated Decay (NMD): NMD is a pathway that degrades mRNAs containing premature stop codons. This mechanism prevents the translation of truncated and potentially harmful proteins.
-
Non-stop Decay (NSD): NSD targets mRNAs lacking stop codons.
These degradation pathways are crucial for eliminating faulty or unwanted transcripts, preventing the production of non-functional or potentially harmful proteins.
The Interplay of Modifications: A Coordinated Effort
The modifications discussed above do not operate in isolation; they are intricately interconnected and often influence each other. For example, the 5' cap and poly(A) tail act synergistically to enhance mRNA stability and translation efficiency. Similarly, RNA methylation patterns can affect splicing efficiency, influencing the final protein product. This complex interplay underscores the sophisticated regulatory mechanisms governing eukaryotic gene expression.
The Biological Significance of Post-Transcriptional Modifications
The biological significance of post-transcriptional modifications extends far beyond simply producing functional proteins. These modifications play crucial roles in:
-
Development and Differentiation: Alternative splicing and RNA editing are particularly important during development, generating the diverse range of proteins needed for cell differentiation and tissue formation.
-
Immune Response: Post-transcriptional modifications are heavily involved in the immune response, regulating the expression of immune-related genes.
-
Disease Pathogenesis: Dysregulation of post-transcriptional modifications is implicated in various diseases, including cancer, neurodegenerative disorders, and infectious diseases. Understanding these modifications is crucial for developing targeted therapies.
Technological Advances and Future Directions
Technological advances have significantly enhanced our understanding of post-transcriptional modifications. High-throughput sequencing technologies, coupled with bioinformatics tools, allow for the comprehensive analysis of transcriptomes, revealing the complex landscape of RNA modifications and their dynamic regulation. Future research will focus on:
-
Uncovering Novel Modifications: New RNA modifications are continually being discovered, expanding our understanding of gene expression regulation.
-
Developing Targeted Therapies: Manipulating post-transcriptional modifications offers exciting therapeutic possibilities for various diseases.
-
Exploring the Interplay of Modifications: Further investigation into the complex interplay between different modifications is crucial for a complete understanding of their regulatory roles.
In conclusion, the post-transcriptional modifications of eukaryotic transcripts are a multifaceted and essential aspect of gene expression. Their intricate network of interactions orchestrates a precise control over protein synthesis, contributing significantly to cellular function, development, and disease pathogenesis. Continued research into these modifications will undoubtedly unveil further insights into the complexity and elegance of eukaryotic gene regulation and pave the way for new therapeutic interventions.
Latest Posts
Latest Posts
-
Key Distinctions Between Poverty And Neglect
Mar 28, 2025
-
What Is The Purpose Of The Checksum Tcp Field
Mar 28, 2025
-
Select All Vectors With An X Component Of Zero
Mar 28, 2025
-
The Government Of Balearic Is Concerned About A Recession
Mar 28, 2025
-
Suppose Your Expectations Regarding The Stock Market Are As Follows
Mar 28, 2025
Related Post
Thank you for visiting our website which covers about Choose All Modifications Of Eukaryotic Transcripts. . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
