Which Of The Following Would Be Transcribed Into Mrna
Holbox
Mar 23, 2025 · 6 min read
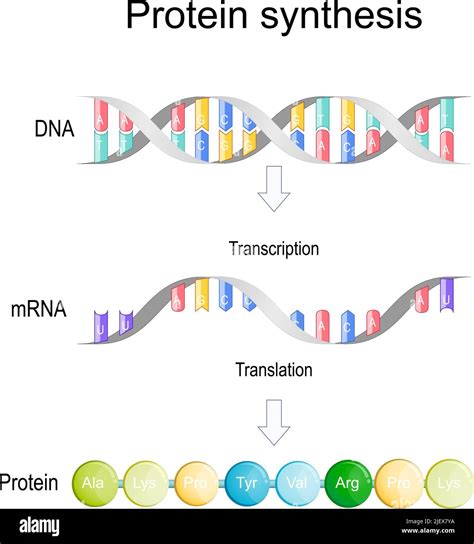
Table of Contents
- Which Of The Following Would Be Transcribed Into Mrna
- Table of Contents
- Which of the Following Would Be Transcribed into mRNA? A Deep Dive into Transcription and Gene Expression
- The Central Dogma: DNA to RNA to Protein
- What is Transcription?
- Identifying Sequences Transcribed into mRNA: A Closer Look
- Examples and Illustrations:
- Practical Application and Future Directions
- Latest Posts
- Latest Posts
- Related Post
Which of the Following Would Be Transcribed into mRNA? A Deep Dive into Transcription and Gene Expression
Understanding which sequences are transcribed into mRNA is fundamental to comprehending gene expression and the central dogma of molecular biology. This article will explore the intricacies of transcription, focusing on identifying DNA sequences that serve as templates for mRNA synthesis. We'll delve into the mechanisms, key players, and factors influencing this crucial process. By the end, you will be well-equipped to determine which DNA sequences are destined for mRNA transcription.
The Central Dogma: DNA to RNA to Protein
The central dogma of molecular biology outlines the flow of genetic information: DNA is transcribed into RNA, which is then translated into protein. This process is essential for life, dictating the synthesis of all proteins necessary for cellular function. However, not all DNA sequences are transcribed; only specific regions, known as genes, undergo transcription.
What is Transcription?
Transcription is the first step in gene expression, where the information encoded in a DNA sequence is copied into a messenger RNA (mRNA) molecule. This process involves several key components:
-
DNA Template: The DNA double helix serves as the template, with only one strand (the template strand or antisense strand) being transcribed. The other strand (the coding strand or sense strand) has a sequence almost identical to the mRNA, except for uracil (U) replacing thymine (T).
-
RNA Polymerase: This enzyme is responsible for synthesizing the mRNA molecule. It binds to the DNA template and adds complementary RNA nucleotides, following the base-pairing rules (A with U, T with A, G with C, and C with G).
-
Promoter Region: A specific DNA sequence upstream of the gene's coding region. It acts as a binding site for RNA polymerase, initiating transcription. The promoter's strength determines the rate of transcription. Different promoters have varying strengths and are recognized by different RNA polymerase isoforms.
-
Transcription Factors: Proteins that bind to specific DNA sequences (including the promoter) to regulate the initiation and rate of transcription. They can either enhance or repress transcription, contributing to the precise control of gene expression.
-
Terminator Region: A DNA sequence that signals the end of transcription. Once the RNA polymerase reaches the terminator, it detaches from the DNA and releases the newly synthesized mRNA molecule.
Identifying Sequences Transcribed into mRNA: A Closer Look
To determine if a given sequence will be transcribed into mRNA, several key aspects must be considered:
1. The Presence of a Promoter:
The most critical factor is the presence of a functional promoter region upstream of the sequence. Without a promoter, RNA polymerase cannot initiate transcription. Promoter sequences are highly conserved and often contain specific consensus sequences (e.g., the TATA box in eukaryotes). The presence and strength of these elements are crucial in determining transcription levels.
2. The Sequence Context:
The surrounding DNA sequence also plays a crucial role. Enhancers and silencers, located further upstream or downstream of the gene, can significantly influence transcription rates by interacting with transcription factors. These regulatory elements can modulate the accessibility of the promoter to RNA polymerase. The presence and position of CpG islands, regions rich in cytosine and guanine nucleotides, also impact gene expression. Methylation of these islands can alter transcription levels.
3. The Type of Sequence:
Not all DNA sequences code for proteins. Some sequences are transcribed into non-coding RNAs (ncRNAs), including ribosomal RNA (rRNA), transfer RNA (tRNA), and microRNAs (miRNAs), each with distinct roles in cellular processes. Introns, non-coding sequences within genes, are transcribed but spliced out before translation. Exons, coding sequences, are spliced together to form the mature mRNA. Therefore, only exonic sequences are ultimately translated into proteins. Understanding whether a sequence resides within an exon or an intron is critical.
4. Chromatin Structure:
The packaging of DNA into chromatin also influences transcription. Tightly packed chromatin (heterochromatin) is generally inaccessible to RNA polymerase, while loosely packed chromatin (euchromatin) allows for easier transcription. Modifications to histone proteins, components of chromatin, can alter chromatin structure and affect transcription.
5. Epigenetic Modifications:
Epigenetic modifications, such as DNA methylation and histone modifications, can alter gene expression without changing the DNA sequence itself. These modifications can affect the accessibility of DNA to RNA polymerase and transcription factors, thus impacting transcription.
Examples and Illustrations:
Let's analyze some hypothetical scenarios:
Scenario 1: A DNA sequence containing a strong promoter region followed by a long stretch of exonic sequences will likely be efficiently transcribed into mRNA, leading to high protein production.
Scenario 2: A DNA sequence with a weak or mutated promoter will show significantly reduced transcription, even if it contains exons. This can lead to low or no protein production.
Scenario 3: A DNA sequence containing a functional promoter but situated within a highly condensed heterochromatin region will likely exhibit low or no transcription. The inaccessibility of the DNA hinders RNA polymerase binding.
Scenario 4: A DNA sequence residing within an intron will be transcribed into pre-mRNA but subsequently spliced out before translation, not becoming part of the mature mRNA.
Scenario 5: A DNA sequence containing a strong promoter followed by intronic sequences and exonic sequences will be transcribed initially, but the intronic regions are then removed through splicing, resulting in a mature mRNA containing only the exonic sequences.
Practical Application and Future Directions
Understanding which sequences are transcribed into mRNA has numerous applications in various fields:
-
Gene Therapy: Precise control over gene expression is crucial for successful gene therapy. Understanding promoter regions and regulatory elements allows scientists to design vectors that efficiently express therapeutic genes.
-
Drug Discovery: Many drugs target specific genes or their mRNA products. Knowing which sequences are transcribed is critical for designing drugs that effectively modulate gene expression.
-
Diagnostics: Measuring mRNA levels is vital in diagnosing various diseases, as changes in mRNA levels often reflect changes in gene expression.
-
Biotechnology: The ability to control transcription is essential for producing recombinant proteins. Understanding the molecular mechanisms of transcription allows scientists to optimize the production of valuable biomolecules.
Ongoing research continues to unveil the complexities of transcription regulation. Advances in next-generation sequencing technologies are providing deeper insights into the transcriptome, the complete set of RNA transcripts in a cell. These studies are expanding our understanding of gene expression and the role of regulatory elements in controlling transcription. The field of epigenetics, investigating heritable changes in gene expression without altering the DNA sequence, is also rapidly progressing. Future research promises to enhance our ability to predict and manipulate transcription, with broad implications across various fields of science and medicine.
This detailed exploration emphasizes the importance of considering multiple factors to determine which DNA sequences are transcribed into mRNA. It's a complex process governed by the interplay of promoter strength, regulatory elements, chromatin structure, epigenetic modifications, and the type of sequence itself. A complete understanding of these factors is crucial for comprehending the intricate mechanisms that drive gene expression and its implications for cellular function, health, and disease.
Latest Posts
Latest Posts
-
A Nation Can Achieve Higher Economic Growth If
Mar 26, 2025
-
Experiment 3 Osmosis Direction And Concentration Gradients
Mar 26, 2025
-
The Hootsuite Bulk Composer Enables You To
Mar 26, 2025
-
A 30 Year Old Woman With A History Of Alcoholism
Mar 26, 2025
-
The Term Deviance Can Be Defined As
Mar 26, 2025
Related Post
Thank you for visiting our website which covers about Which Of The Following Would Be Transcribed Into Mrna . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
