Which Of The Events Occur During Eukaryotic Translation Initiation
Holbox
Mar 16, 2025 · 6 min read
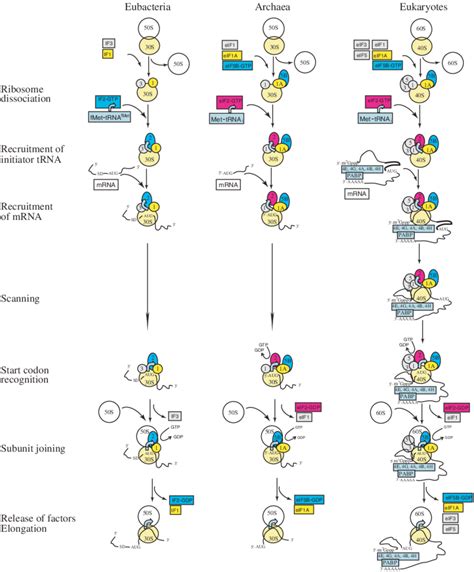
Table of Contents
Eukaryotic Translation Initiation: A Deep Dive into the Complex Process
Eukaryotic translation initiation, the first step in protein synthesis, is a remarkably intricate process involving numerous initiation factors (eIFs), ribosome subunits, mRNA, and initiator tRNA. Unlike its prokaryotic counterpart, eukaryotic initiation is significantly more complex, reflecting the greater structural complexity of eukaryotic cells and the need for precise regulation. Understanding this intricate dance of molecular players is crucial for grasping the fundamental mechanisms of gene expression and cellular regulation. This article delves into the key events of eukaryotic translation initiation, providing a comprehensive overview of the process.
The Players: Key Components of Eukaryotic Translation Initiation
Before diving into the steps, let's introduce the key players:
-
mRNA (messenger RNA): Carries the genetic code from DNA to the ribosome. It contains the start codon (AUG) and the coding sequence. Key features like the 5' cap and 3' poly(A) tail play crucial roles in initiation.
-
Ribosomal Subunits: The eukaryotic ribosome consists of a 40S small subunit and a 60S large subunit. These subunits come together during initiation to form the 80S ribosome, the protein synthesis machinery.
-
Initiator tRNA (methionine tRNA, tRNAiMet): Carries the initiator methionine amino acid, which is the first amino acid in most eukaryotic proteins. This is distinct from the tRNA that carries methionine during elongation.
-
eIFs (eukaryotic Initiation Factors): A large family of proteins that orchestrate the various steps of initiation. Each eIF plays a specific role, and their interactions are tightly regulated. Key eIFs discussed later include eIF1, eIF1A, eIF2, eIF2B, eIF3, eIF4A, eIF4B, eIF4E, eIF4G, eIF5, and eIF5B.
-
GTP (Guanosine Triphosphate): Provides energy for several steps in the initiation process.
Stages of Eukaryotic Translation Initiation: A Step-by-Step Guide
Eukaryotic translation initiation can be broadly categorized into several key steps:
1. Pre-initiation Complex Formation: Preparing the Small Ribosomal Subunit
This initial phase involves the assembly of the 43S pre-initiation complex (PIC). This complex is fundamental for the entire process and comprises the following crucial steps:
-
eIF2-GTP-tRNAiMet ternary complex formation: eIF2, a GTPase, binds to GTP and tRNAiMet, forming a ternary complex. This complex is crucial for delivering the initiator tRNA to the ribosome.
-
40S ribosomal subunit recruitment: The 40S ribosomal subunit binds to eIF3 and other eIFs (e.g., eIF1, eIF1A), forming a complex ready to receive the ternary complex. eIF3 plays a crucial role in preventing premature 60S subunit joining.
-
Joining of the ternary complex: The eIF2-GTP-tRNAiMet ternary complex joins the 40S subunit complex, completing the 43S pre-initiation complex. This complex is now ready to scan the mRNA.
2. mRNA Activation and Recruitment: Getting the Message Ready
Before the ribosome can begin translation, the mRNA must be prepared:
-
5' cap recognition: eIF4E, a cap-binding protein, binds to the 5' methylguanosine cap of the mRNA. This is the initial step in mRNA recruitment. This interaction is critical for the circularization of the mRNA.
-
eIF4A and eIF4B recruitment: These helicase proteins unwind secondary structures within the 5' untranslated region (UTR) of the mRNA, facilitating ribosomal scanning.
-
eIF4G interaction: eIF4G acts as a scaffold, connecting eIF4E (bound to the 5' cap) to other initiation factors and the poly(A)-binding protein (PABP) bound to the 3' poly(A) tail. This interaction leads to mRNA circularization, enhancing translation efficiency by creating a closed-loop structure. This increases the stability of the mRNA and promotes re-initiation after completion of one round of translation.
-
Recruitment of the 43S PIC to the mRNA: The 43S pre-initiation complex is recruited to the mRNA through interactions between eIF4G and other eIFs.
3. mRNA Scanning and Start Codon Recognition: Finding the Starting Point
This crucial step involves the scanning of the mRNA by the 43S PIC:
-
Ribosome scanning: The 43S PIC scans the 5' UTR of the mRNA in a 5' to 3' direction until it encounters the start codon (AUG). This scanning is an ATP-dependent process, utilizing the energy provided by ATP hydrolysis. The scanning process is aided by the helicase activity of eIF4A and eIF4B.
-
Start codon recognition: Once the 43S PIC reaches the AUG start codon, a conformational change occurs. This is facilitated by the interaction of the initiator tRNA anticodon with the AUG codon. The surrounding Kozak sequence (consensus sequence surrounding the start codon – GCCRCCAUGG) also contributes to start codon recognition.
4. 60S Subunit Joining and Initiation Complex Formation: Building the Translation Machinery
This stage culminates in the formation of the complete 80S initiation complex:
-
GTP hydrolysis by eIF2: eIF2 hydrolyzes its bound GTP, triggering conformational changes that release eIFs from the complex, particularly eIF2 and eIF5.
-
Recruitment of eIF5B-GTP: eIF5B, another GTPase, binds to the complex. This binding is critical for the joining of the 60S subunit.
-
60S subunit joining: The 60S ribosomal subunit joins the complex, aided by eIF5B. This forms the complete 80S initiation complex.
-
GTP hydrolysis by eIF5B: eIF5B hydrolyzes its bound GTP, releasing itself from the complex. The 80S ribosome is now ready to initiate elongation.
-
Positioning of initiator tRNA in the P site: The initiator tRNA (tRNAiMet) is positioned in the peptidyl (P) site of the ribosome, ready to accept the next amino acid.
Regulation of Eukaryotic Translation Initiation: Fine-Tuning Protein Synthesis
The initiation phase is a heavily regulated process. Several mechanisms contribute to this regulation:
-
Phosphorylation of eIF2: Phosphorylation of eIF2 inhibits its ability to exchange GDP for GTP, effectively halting translation initiation. This is a crucial mechanism in response to stress conditions such as amino acid starvation or viral infection.
-
Phosphorylation of other eIFs: Phosphorylation of other eIFs, such as eIF4E, can also modulate translation initiation rates.
-
Regulation of eIF4E availability: The availability of eIF4E is tightly controlled, affecting the overall rate of translation.
-
Upstream Open Reading Frames (uORFs): These small open reading frames in the 5' UTR can influence the efficiency of translation initiation of the main coding sequence. They can either promote or inhibit translation depending on their context.
-
RNA secondary structure: The presence of secondary structure within the 5' UTR can hinder ribosome scanning and thus affect translation initiation.
-
MicroRNAs (miRNAs): These small RNA molecules can bind to specific mRNAs, affecting their translation efficiency. They can lead to either translational repression or mRNA degradation.
Conclusion: A Complex Process with Far-Reaching Implications
Eukaryotic translation initiation is a multi-step process involving a complex interplay of numerous initiation factors, ribosomes, and mRNA. The precise regulation of this process is essential for controlling gene expression, responding to cellular stress, and maintaining cellular homeostasis. Dysregulation of translation initiation is implicated in numerous diseases, including cancer and various neurological disorders. Therefore, a comprehensive understanding of this process is not only fundamental to basic biology but also crucial for the development of novel therapeutic strategies. Further research into the intricate mechanisms governing translation initiation continues to unravel its complexities and reveal its significance in health and disease. The detailed mechanisms of initiation factor interactions, conformational changes within the ribosome, and the regulation of each stage remain active areas of investigation. This sophisticated and highly regulated process truly underscores the remarkable efficiency and precision of cellular machinery.
Latest Posts
Latest Posts
-
Which Statement Is True Regarding A Minor Beneficiary
Mar 16, 2025
-
Which Is True Of Inducements In Research
Mar 16, 2025
-
Complete The Balanced Neutralization Equation For The Reaction Below
Mar 16, 2025
-
What Conclusion Can Be Drawn Based On At
Mar 16, 2025
-
Endangering Other People On A Highway Dmv
Mar 16, 2025
Related Post
Thank you for visiting our website which covers about Which Of The Events Occur During Eukaryotic Translation Initiation . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
