What Is Wrong With The Following Piece Of Mrna Taccaggatcactttgcca
Holbox
Mar 15, 2025 · 6 min read
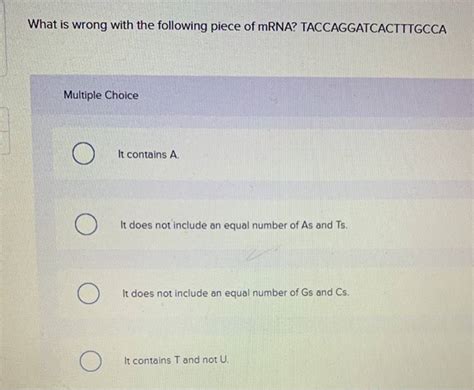
Table of Contents
- What Is Wrong With The Following Piece Of Mrna Taccaggatcactttgcca
- Table of Contents
- What's Wrong with the mRNA Sequence: TACCAAGATCACCTTTGCCA? A Deep Dive into mRNA Structure and Function
- Understanding mRNA: The Messenger Molecule
- Decoding the Sequence: TACCAAGATCACCTTTGCCA
- Codon Usage Bias: A Subtle but Significant Factor
- Potential for Premature Termination Codons
- mRNA Stability and Degradation: A Critical Aspect
- Influence of the 5' and 3' Untranslated Regions (UTRs)
- The Importance of Post-Transcriptional Modifications
- The Context Matters: Cellular Environment and Gene Regulation
- Potential for Misfolding and Aggregation
- Practical Applications and Further Analysis
- Latest Posts
- Latest Posts
- Related Post
What's Wrong with the mRNA Sequence: TACCAAGATCACCTTTGCCA? A Deep Dive into mRNA Structure and Function
The mRNA sequence TACCAAGATCACCTTTGCCA presents several potential issues, depending on the context and intended function. This article will explore these problems in detail, examining aspects of mRNA structure, translation, and potential implications for gene expression. We'll cover topics such as codon usage bias, potential premature termination codons, and the impact of sequence context on mRNA stability and translation efficiency.
Understanding mRNA: The Messenger Molecule
Before diving into the specifics of the given sequence, it's crucial to understand the basic function of messenger RNA (mRNA). mRNA acts as an intermediary between DNA and protein synthesis. It's a single-stranded RNA molecule transcribed from a DNA template, carrying the genetic code from the nucleus to the ribosomes in the cytoplasm, where it directs protein synthesis. This process, known as translation, involves the decoding of the mRNA sequence into a specific amino acid sequence, forming a polypeptide chain that folds into a functional protein.
Decoding the Sequence: TACCAAGATCACCTTTGCCA
Let's break down the provided mRNA sequence: TACCAAGATCACCTTTGCCA. Each three-nucleotide sequence, called a codon, codes for a specific amino acid. Using the standard genetic code, we can translate this mRNA sequence:
- TAC: Tyrosine (Tyr or Y)
- CAA: Glutamine (Gln or Q)
- GAT: Aspartic acid (Asp or D)
- CAC: Histidine (His or H)
- CTT: Leucine (Leu or L)
- TGC: Cysteine (Cys or C)
- CA: This is not a complete codon.
The incomplete codon at the end, "CA," highlights the first potential problem. mRNA sequences must be multiples of three nucleotides to ensure proper codon reading frame. An incomplete codon disrupts this frame, leading to a truncated translation and potentially a non-functional protein.
Codon Usage Bias: A Subtle but Significant Factor
Even if the sequence were complete (let's assume an additional base, making it TACCAAGATCACCTTTGCCA), we need to consider codon usage bias. Codon usage bias refers to the non-random preference for certain codons over others, even though they may code for the same amino acid. This bias varies significantly across different organisms and even within different genes of the same organism.
The specific codons used can influence translation efficiency. Codons frequently used in an organism's genes are typically translated more quickly and accurately due to the higher abundance of corresponding tRNAs (transfer RNAs). Conversely, infrequently used codons can lead to slower translation and even ribosome stalling. Without knowing the organism of origin, it's impossible to determine the degree to which this sequence adheres to the optimal codon usage bias. An analysis comparing the codon usage of this sequence to that of the organism's typical gene sequences would provide valuable insight.
Potential for Premature Termination Codons
The sequence, even if completed, does not contain any obvious premature stop codons (UAA, UAG, UGA). However, the context and surrounding sequences can play a significant role. A sequence with potential for secondary structures like hairpin loops could mask or create apparent stop codons through frameshifting. Analyzing the secondary structure prediction of the mRNA sequence would help determine if any such structures are present.
mRNA Stability and Degradation: A Critical Aspect
mRNA stability is another crucial factor affecting protein production. mRNA molecules are constantly subjected to degradation by cellular enzymes called RNases. Certain mRNA sequences are more prone to degradation than others. The sequence TACCAAGATCACCTTTGCCA does not immediately reveal any obvious instability signals, such as AU-rich elements (AREs), which are often found in the 3' untranslated region (3'UTR) of unstable mRNAs. However, a full analysis, including the 5' and 3' untranslated regions and any potential secondary structures, is necessary for a comprehensive assessment.
Influence of the 5' and 3' Untranslated Regions (UTRs)
The sequence provided only shows the coding sequence. The 5' and 3' UTRs are critical for mRNA stability, translation efficiency, and localization. The 5' UTR contains the ribosome binding site (RBS), also known as the Shine-Dalgarno sequence in prokaryotes, crucial for initiating translation. The 3' UTR plays a role in mRNA stability, polyadenylation, and translational regulation. Without information about the UTRs, we cannot fully assess the functionality of the mRNA.
The Importance of Post-Transcriptional Modifications
Even if the coding sequence is perfectly fine, post-transcriptional modifications can significantly impact mRNA functionality. These modifications, including splicing, capping, and polyadenylation, are essential for mRNA processing, stability, and translation. The provided sequence offers no information about these critical modifications. For instance, incorrect splicing can lead to frameshifts, premature termination, or the inclusion of non-coding sequences, ultimately disrupting protein synthesis.
The Context Matters: Cellular Environment and Gene Regulation
mRNA function is heavily influenced by the cellular environment and complex gene regulatory networks. The sequence alone does not provide information about the cellular context in which it operates. Factors such as transcription factors, RNA-binding proteins, and microRNAs can interact with the mRNA, affecting its stability, translation, and overall functionality. This interaction is complex and often involves intricate feedback loops and regulatory mechanisms.
Potential for Misfolding and Aggregation
The resulting protein from translation (assuming a complete and correct sequence) could exhibit misfolding and aggregation problems. These issues are not readily apparent from the mRNA sequence alone, but the amino acid sequence derived from the translation is a major contributing factor. Certain amino acid sequences are prone to forming beta-sheets, which can lead to misfolding and aggregation, potentially causing cellular damage.
Practical Applications and Further Analysis
To fully understand the "wrongness" of the mRNA sequence TACCAAGATCACCTTTGCCA, further investigation is needed. This includes:
- Completing the sequence: Determining the correct length and adding any missing nucleotides is crucial for accurate translation.
- Determining the organism of origin: This allows for analysis of codon usage bias, relevant to translational efficiency.
- Analyzing the 5' and 3' UTRs: These regions are crucial for mRNA stability, translation initiation, and other regulatory processes.
- Investigating post-transcriptional modifications: Splicing, capping, and polyadenylation are all vital for correct mRNA processing.
- Studying the secondary structure: Predicting the mRNA secondary structure can reveal potential hairpin loops or other structures that might disrupt translation.
- Analyzing the protein product (if any): Determining the amino acid sequence, predicting its structure, and investigating potential misfolding or aggregation issues are essential.
- Considering the cellular context: Understanding the cellular environment and regulatory networks is crucial for comprehending the sequence's functional context.
In conclusion, simply stating what's "wrong" with the mRNA sequence TACCAAGATCACCTTTGCCA is insufficient without considering the broader context. The incomplete codon is a clear issue, but further analysis is necessary to address potential issues related to codon usage bias, mRNA stability, translation efficiency, post-transcriptional modifications, and the overall cellular environment. A comprehensive analysis incorporating bioinformatic tools and experimental validation is essential for a thorough understanding of this sequence's functional properties or lack thereof.
Latest Posts
Latest Posts
-
If Asked To Analyze The Structures Of A Cadaver
Mar 16, 2025
-
A Manager Evaluates A Suboriated Job Perfomrance And
Mar 16, 2025
-
In The Molecule Bri Which Atom Is The Negative Pole
Mar 16, 2025
-
Being Civilly Liable Means A Server Or Seller Of Alcohol
Mar 16, 2025
-
What Does The Function Labeled Continue From Previous Section Do
Mar 16, 2025
Related Post
Thank you for visiting our website which covers about What Is Wrong With The Following Piece Of Mrna Taccaggatcactttgcca . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
