Rna Polymerase Is Guided By The
Holbox
Mar 21, 2025 · 6 min read
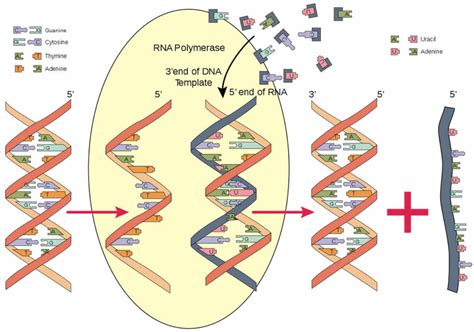
Table of Contents
- Rna Polymerase Is Guided By The
- Table of Contents
- RNA Polymerase is Guided By: A Deep Dive into Transcription Initiation and Regulation
- The Promoters: The Starting Point for Transcription
- Bacterial Promoters: A Simpler System
- Eukaryotic Promoters: A More Complex Orchestration
- RNA Polymerase II: The Workhorse of Eukaryotic Transcription
- Chromatin Structure: A Regulatory Barrier
- Histone Modifications: The Epigenetic Landscape
- Chromatin Remodeling Complexes: Restructuring the Chromatin
- Post-Transcriptional Regulation: Fine-tuning Gene Expression
- Conclusion: A Multifaceted Guidance System
- Latest Posts
- Latest Posts
- Related Post
RNA Polymerase is Guided By: A Deep Dive into Transcription Initiation and Regulation
RNA polymerase, the enzyme responsible for transcription, isn't just a lone wolf wandering through the genome. Its journey, the process of creating RNA from a DNA template, is intricately guided and regulated. Understanding how RNA polymerase is guided is crucial to understanding gene expression, a fundamental process of life. This comprehensive article will explore the multifaceted mechanisms that direct RNA polymerase to specific genes, control the initiation of transcription, and ultimately determine which proteins are synthesized within a cell.
The Promoters: The Starting Point for Transcription
The journey begins at the promoter, a specific DNA sequence located upstream of the gene. The promoter acts as a landing pad, signaling where RNA polymerase should bind and initiate transcription. Promoters aren't uniform; their sequences vary, leading to differences in the strength and efficiency of transcription initiation. The key players in promoter recognition are transcription factors, proteins that bind to specific DNA sequences within the promoter region.
Bacterial Promoters: A Simpler System
Bacterial promoters typically contain two crucial elements: the -10 sequence (Pribnow box) and the -35 sequence. These sequences are named for their position relative to the transcription start site (+1), which is the first nucleotide transcribed into RNA. The sigma factor, a subunit of bacterial RNA polymerase, recognizes and binds to these sequences. Different sigma factors have different preferences for promoter sequences, enabling the bacterium to regulate the expression of different sets of genes in response to environmental changes. Strong promoters exhibit sequences highly similar to the consensus sequences, while weak promoters display greater variation. This difference in sequence dictates the frequency of transcription initiation.
Eukaryotic Promoters: A More Complex Orchestration
Eukaryotic promoters are significantly more complex than their bacterial counterparts. While they also include core promoter elements like the TATA box (analogous to the -10 sequence), they also incorporate a vast array of other regulatory sequences, including promoter-proximal elements and enhancers. These elements act as binding sites for a multitude of transcription factors, creating a highly intricate regulatory network.
-
General Transcription Factors (GTFs): These are essential proteins that assemble at the promoter, forming the pre-initiation complex (PIC). They are required for the recruitment and positioning of RNA polymerase II (the polymerase responsible for transcribing most protein-coding genes). Key GTFs include TFIID (which binds the TATA box), TFIIB, TFIIF, TFIIE, and TFIIH.
-
Promoter-Proximal Elements: These sequences are located closer to the transcription start site than enhancers and typically bind specific transcription factors that either activate or repress transcription. Their effects are often localized to nearby genes.
-
Enhancers: Enhancers are cis-regulatory elements that can be located thousands of base pairs upstream or downstream of the gene they regulate. They enhance transcription by binding activator proteins, which interact with the PIC through looping of the DNA. This interaction allows for long-range gene regulation.
-
Silencers: In contrast to enhancers, silencers are cis-regulatory elements that repress transcription. They bind repressor proteins, inhibiting the formation or activity of the PIC.
The interplay between these various promoter elements and their associated transcription factors determines the precise level of gene expression. This intricate system enables the cell to fine-tune gene expression in response to internal and external signals.
RNA Polymerase II: The Workhorse of Eukaryotic Transcription
In eukaryotes, RNA polymerase II is responsible for the transcription of the vast majority of protein-coding genes. Its recruitment to the promoter is a complex process, requiring the precise assembly of the pre-initiation complex (PIC). The PIC formation involves several steps:
-
TFIID binding: TFIID, a complex containing the TATA-binding protein (TBP) and TBP-associated factors (TAFs), recognizes and binds to the TATA box or other core promoter elements.
-
Recruitment of other GTFs: The binding of TFIID recruits other general transcription factors, including TFIIB, TFIIF, TFIIE, and TFIIH.
-
RNA Polymerase II recruitment: TFIIF plays a crucial role in recruiting RNA polymerase II to the PIC.
-
Formation of the open complex: TFIIH possesses helicase activity, which unwinds the DNA at the transcription start site, creating the transcription bubble.
-
Initiation of transcription: After the formation of the open complex, RNA polymerase II initiates transcription, synthesizing the nascent RNA molecule.
This highly orchestrated process ensures that transcription is initiated accurately and efficiently.
Chromatin Structure: A Regulatory Barrier
The DNA in eukaryotic cells isn't simply a naked molecule; it's packaged into chromatin, a complex structure composed of DNA and histone proteins. Chromatin structure plays a critical role in regulating gene expression. Tightly packed chromatin (heterochromatin) is transcriptionally inactive, while loosely packed chromatin (euchromatin) is accessible to RNA polymerase and transcription factors.
Histone Modifications: The Epigenetic Landscape
Histones can be modified by various chemical groups, such as acetyl, methyl, and phosphate groups. These modifications alter the chromatin structure, influencing the accessibility of DNA to RNA polymerase and transcription factors. For example, histone acetylation generally leads to chromatin relaxation and increased transcription, while histone methylation can either activate or repress transcription, depending on the specific residue and the number of methyl groups added.
Chromatin Remodeling Complexes: Restructuring the Chromatin
Chromatin remodeling complexes are large protein complexes that use ATP hydrolysis to alter the position and structure of nucleosomes, the fundamental units of chromatin. These complexes can either slide nucleosomes along the DNA, evict nucleosomes, or replace histone variants, making DNA more or less accessible to transcription machinery. This dynamic remodeling of chromatin plays a crucial role in regulating gene expression.
Post-Transcriptional Regulation: Fine-tuning Gene Expression
While the initiation of transcription is a critical control point, gene expression is also regulated post-transcriptionally. This involves processes that affect the stability, processing, and translation of the RNA transcript. These processes can fine-tune gene expression in response to changing cellular conditions.
-
RNA processing: Eukaryotic pre-mRNA transcripts undergo various processing steps, including capping, splicing, and polyadenylation. These processes are crucial for mRNA stability and translation efficiency. Alterations in these processes can significantly impact gene expression levels.
-
RNA stability: The stability of mRNA molecules is another crucial control point. mRNAs have varying half-lives, and their degradation rates can be influenced by various factors, including the presence of specific sequences or the activity of RNA-binding proteins.
-
RNA interference (RNAi): RNAi is a powerful mechanism that silences gene expression by targeting specific mRNAs for degradation or translational repression. Small interfering RNAs (siRNAs) and microRNAs (miRNAs) are key players in this pathway.
Conclusion: A Multifaceted Guidance System
The guidance of RNA polymerase is a complex and tightly regulated process, involving a multitude of factors and mechanisms. From the recognition of promoter sequences to the dynamic remodeling of chromatin and post-transcriptional regulation, each step contributes to the precise control of gene expression. Understanding these mechanisms is crucial for comprehending the intricacies of cellular processes, development, and disease. Further research into the intricate interactions between RNA polymerase, transcription factors, chromatin structure, and post-transcriptional regulation will undoubtedly continue to reveal new layers of complexity and offer novel insights into the fundamental processes of life. The journey of RNA polymerase, far from being a simple linear path, is a dynamic and highly orchestrated dance that defines the very essence of cellular life.
Latest Posts
Latest Posts
-
The Primary Organizational Buying Objective For Business Firms Is To
Mar 28, 2025
-
A Statement Of Comprehensive Income Does Not Include
Mar 28, 2025
-
Use The Following Cell Phone Airport Data
Mar 28, 2025
-
Slack Is An Example Of Collaboration Software True Or False
Mar 28, 2025
-
If A Person Is Severely Dehydrated Their Extracellular
Mar 28, 2025
Related Post
Thank you for visiting our website which covers about Rna Polymerase Is Guided By The . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
