How Can Dna Methylation Silence Transcription
Holbox
Mar 29, 2025 · 6 min read
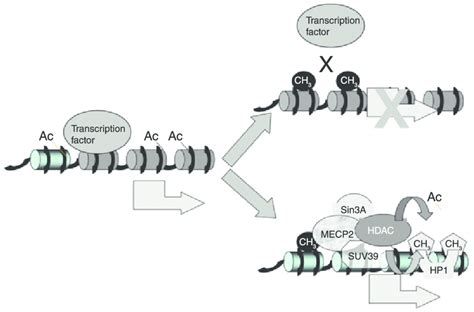
Table of Contents
- How Can Dna Methylation Silence Transcription
- Table of Contents
- How DNA Methylation Silences Transcription: A Deep Dive
- The Mechanisms Behind Methylation-Mediated Transcriptional Silencing
- 1. Direct Interference with Transcription Factor Binding
- 2. Recruitment of Methyl-CpG-Binding Proteins (MBDs)
- 3. Histone Modification and Chromatin Remodeling
- 4. Chromatin Compaction and Formation of Heterochromatin
- CpG Islands and the Regulation of Gene Expression
- The Dynamic Nature of DNA Methylation
- Implications of DNA Methylation in Health and Disease
- Conclusion
- Latest Posts
- Latest Posts
- Related Post
How DNA Methylation Silences Transcription: A Deep Dive
DNA methylation, a fundamental epigenetic modification, plays a crucial role in regulating gene expression. It's a process where a methyl group (CH₃) is added to a cytosine base, typically within CpG dinucleotides (a cytosine base followed by a guanine base). This seemingly simple chemical addition can have profound consequences, often leading to transcriptional silencing. Understanding how DNA methylation achieves this silencing is vital to comprehending various biological processes, including development, disease, and aging.
The Mechanisms Behind Methylation-Mediated Transcriptional Silencing
The silencing effect of DNA methylation isn't a direct, single-step process. Instead, it operates through a complex interplay of mechanisms that ultimately prevent gene transcription. Here's a detailed breakdown:
1. Direct Interference with Transcription Factor Binding
The addition of a methyl group to a cytosine base can physically impede the binding of transcription factors (TFs) to their cognate DNA sequences. Many transcription factors require specific interactions with the DNA's base pairs to bind effectively. Methylation alters the chemical structure of the DNA, creating steric hindrance and preventing these crucial interactions. This is particularly relevant for TFs that recognize CpG sites within their binding motifs.
Example: Specific transcription factors, such as those containing a CxxC domain (characterized by a cysteine-rich motif), are known to have a preference for unmethylated CpG sites. Methylation of these sites can effectively prevent their binding, thus blocking the initiation of transcription.
2. Recruitment of Methyl-CpG-Binding Proteins (MBDs)
Methyl-CpG-binding proteins (MBDs) are a family of proteins that specifically recognize and bind to methylated CpG sites. This binding acts as a crucial step in establishing a repressive chromatin environment. MBDs themselves don't directly silence transcription, but they serve as scaffolding proteins, recruiting other chromatin-modifying complexes to the methylated region.
Key MBD proteins and their roles:
- MeCP2: One of the most extensively studied MBDs, MeCP2 is involved in various aspects of transcriptional repression. It recruits histone deacetylases (HDACs) and other chromatin-remodeling complexes.
- MBD1, MBD2, MBD3, MBD4: These proteins also bind to methylated CpG sites and participate in gene silencing through various mechanisms, often involving interactions with other repressive factors.
3. Histone Modification and Chromatin Remodeling
The recruitment of MBDs often leads to alterations in histone modifications, a major aspect of chromatin remodeling. Histones, proteins around which DNA is wrapped, undergo various post-translational modifications, such as acetylation, methylation, and phosphorylation. These modifications influence the accessibility of DNA to the transcriptional machinery.
How MBDs alter histone modifications:
- Histone deacetylation: MBDs often recruit histone deacetylases (HDACs), enzymes that remove acetyl groups from histone tails. Deacetylation leads to a more condensed chromatin structure, making the DNA less accessible to RNA polymerase II and other transcriptional factors, thus silencing gene expression.
- Histone methylation: MBDs can also recruit histone methyltransferases (HMTs), which add methyl groups to histone tails. The specific histone residue and the number of methyl groups added dictate the outcome, which can either be repressive or activating. In the context of gene silencing, MBDs typically recruit HMTs that catalyze repressive histone marks, such as H3K9me3 (trimethylation of lysine 9 on histone H3) or H3K27me3.
4. Chromatin Compaction and Formation of Heterochromatin
The combined effect of histone modifications and the recruitment of chromatin-remodeling complexes leads to a significant compaction of chromatin structure. This compaction forms a heterochromatic state, a highly condensed form of chromatin that is transcriptionally inactive. Heterochromatin is characterized by its dense packing, making the DNA inaccessible to the transcriptional machinery.
The role of HP1 proteins: Heterochromatin protein 1 (HP1) proteins are essential for maintaining the heterochromatic state. They are recruited to regions with specific histone modifications, such as H3K9me3, and help to consolidate the compacted chromatin structure, further reinforcing transcriptional silencing.
CpG Islands and the Regulation of Gene Expression
CpG islands are regions of DNA with a high density of CpG dinucleotides. They are often located in the promoter regions of genes. The methylation status of CpG islands is closely correlated with gene expression.
Methylation of CpG islands typically leads to gene silencing. This is because the methylation of CpG sites within promoter regions disrupts the binding of activating transcription factors and recruits MBDs, initiating the cascade of events described above. Conversely, unmethylated CpG islands are associated with active gene transcription.
The Dynamic Nature of DNA Methylation
It's crucial to understand that DNA methylation isn't a static, irreversible process. It's a dynamic modification, meaning it can be established, maintained, or erased throughout the lifespan of an organism. This dynamic nature is essential for development, cellular differentiation, and response to environmental stimuli.
DNA methyltransferases (DNMTs) and Demethylases:
- DNMTs: These enzymes are responsible for establishing and maintaining DNA methylation patterns. DNMT1 maintains pre-existing methylation patterns during DNA replication, ensuring that methylation patterns are passed on to daughter cells. DNMT3A and DNMT3B establish de novo methylation.
- Ten-eleven translocation (TET) enzymes: These enzymes are crucial for DNA demethylation. They catalyze the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), initiating a pathway that eventually leads to the removal of the methyl group.
The interplay between DNMTs and TET enzymes allows for a precise regulation of DNA methylation patterns, which is crucial for the dynamic control of gene expression.
Implications of DNA Methylation in Health and Disease
Dysregulation of DNA methylation plays a significant role in various diseases, including cancer. Aberrant methylation patterns are frequently observed in cancer cells, contributing to the development and progression of the disease.
Examples of the role of DNA methylation in disease:
- Cancer: Hypermethylation of tumor suppressor genes can lead to their silencing, contributing to uncontrolled cell growth and cancer development. Conversely, hypomethylation of oncogenes can lead to their activation, further promoting cancer progression.
- Neurological disorders: Abnormal DNA methylation patterns are implicated in several neurological disorders, such as Rett syndrome and autism spectrum disorder. These alterations affect the expression of genes crucial for brain development and function.
- Cardiovascular disease: Changes in DNA methylation patterns are associated with an increased risk of cardiovascular disease. These changes can affect the expression of genes involved in cholesterol metabolism and inflammation.
Conclusion
DNA methylation is a powerful epigenetic mechanism that can effectively silence transcription. It operates through a multifaceted approach, involving direct interference with transcription factor binding, recruitment of MBDs, histone modifications, chromatin remodeling, and the formation of heterochromatin. The dynamic nature of DNA methylation, regulated by DNMTs and TET enzymes, underscores its essential role in maintaining cellular homeostasis and responding to environmental cues. Dysregulation of this intricate process has far-reaching consequences, contributing to the development and progression of numerous diseases. Future research will continue to unravel the complexities of DNA methylation and its impact on health and disease, paving the way for novel therapeutic strategies. Understanding the intricate mechanisms involved is key to developing targeted therapies that can modulate DNA methylation patterns to treat various diseases.
Latest Posts
Latest Posts
-
What Two Things Does Fair Chase Emphasize
Apr 01, 2025
-
Productivity Growth Is Also Closely Linked To
Apr 01, 2025
-
When Sugar Is Mixed With Water Equilibrium Is Reached When
Apr 01, 2025
-
Check In Check Out Managing Hotel Operations Vallen
Apr 01, 2025
-
A Company Sold A Machine For 15000 In Cash
Apr 01, 2025
Related Post
Thank you for visiting our website which covers about How Can Dna Methylation Silence Transcription . We hope the information provided has been useful to you. Feel free to contact us if you have any questions or need further assistance. See you next time and don't miss to bookmark.
